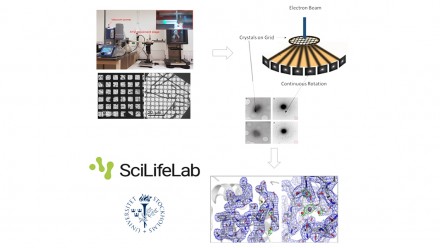
MicroED for Biological Samples: Requirements, Sample Preparation and Data Processing
3D electron diffraction techniques used for decades in material science were recently adapted to study biological samples. Microcrystal electron diffraction (MicroED) enables crystals considered too small (sub-microns) for conventional X-ray diffraction to be used to solve protein structures1-7.
We will present how to:
- choose / grow crystals suitable for electron diffraction
- prepare the sample grids. A newly and easy to implement method for preparing cryo-EM specimens of both single particles and micro-crystals (Preassis8) will be presented.
- diffract these crystals under the electron beam using the (continuous) rotation method2,3
- process the data using standard crystallographic routines9
Examples of applications in biological studies will illustrate the presentation.
1. Shi, D., Nannenga, B. L., Iadanza, M. G. & Gonen, T. Three-dimensional electron crystallography of protein microcrystals. Elife 2, e01345 (2013).
2. Nederlof, I., van Genderen, E., Li, Y. W. & Abrahams, J. P. A Medipix quantum area detector allows rotation electron diffraction data collection from submicrometre three-dimensional protein crystals. Acta Cryst. D 69, 1223–1230 (2013).
3. Nannenga, B. L., Shi, D., Leslie, A. G. W. & Gonen, T. High-resolution structure determination by continuous-rotation data collection in MicroED. Nat. Methods 11, 927–930 (2014).
4. Rodriguez, J. A. et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature 525, 486–490 (2015).
5. Yonekura, K., Kato, K., Ogasawara, M., Tomita, M. & Toyoshima, C. Electron crystallography of ultrathin 3D protein crystals: Atomic model with charges. Proc. Natl. Acad. Sci. U. S. A. 112, 3368–3373 (2015).
6. Clabbers, M. T. B. et al. Protein structure determination by electron diffraction using a single three-dimensional nanocrystal. Acta Cryst. D 73, 738–748 (2017).
7. Xu, H. et al. A Rare Lysozyme Crystal Form Solved Using Highly Redundant Multiple Electron Diffraction Datasets from Micron-Sized Crystals. Structure 26, 667-675.e3 (2018).
8. Zao, J. et al. A simple pressure-assisted method for cryo-EM specimen preparation. Biorxiv.
9. Clabbers, M. T. B., Gruene, T., Parkhurst, J. M., Abrahams, J. P. & Waterman, D. G. Electron diffraction data processing with DIALS. Acta Crystallogr. Sect. D Biol. Crystallogr. 74, 506–518 (2018).
Bio:
Structural biologist combining expertise in Biochemistry (cloning, expression, purification, stabilization and engineering of soluble or membrane proteins, characterization and assay development), Biophysics (Protein crystallography, ITC, SPR, MST...) and Bioinformatics (sequences analysis, de novo and homology modeling, molecular dynamic simulation, small molecule-protein/protein-protein docking),
I have a strong interest in linking
After my thesis in Montreal studying FBP-aldolases of human pathogens, I studied several system where I focused on linking protein dynamic with function and to apply my discoveries into drug-design.