A yeast surface display nanobody factory
Using yeast surface display synthetic biology for a nanobody production platform.
Research themes
Project status
Content navigation
About
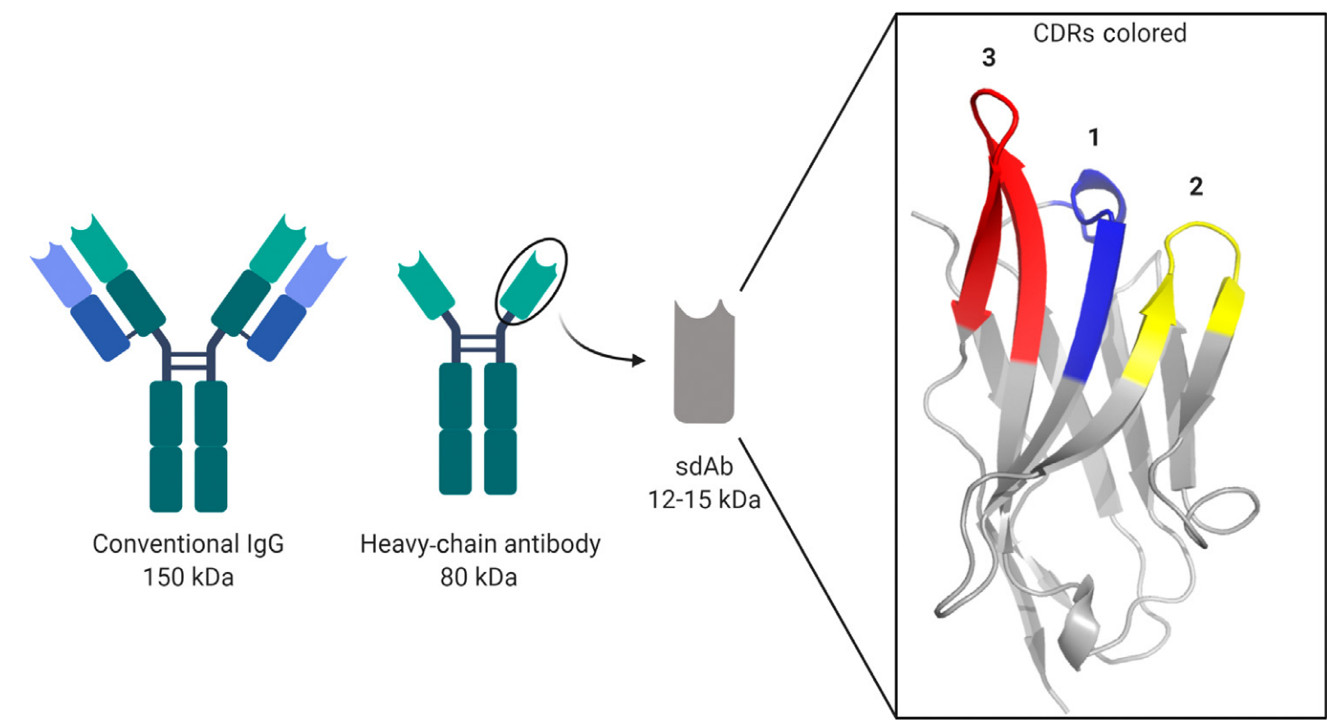
Recently, a protocol for yeast surface display of a synthetic nanobody library was published, allowing the identification of high-affinity nanobodies to a particular antigen, without the immunisation of animals. Nanobodies are derived from the heavy chain antibodies found in the immune system of the Camelid family of mammals, such as llamas and camels, which differ from most other animal derived Immunoglobulin (IgG) antibodies in that they consist of a dimer of Variable Heavy Chain fragments only (Figure 1). Nanobodies are comprised of the complementarity determining domain (VHH) of each chain in isolation, and are therefore a small (12-15 kDa), monomeric alternative to conventional antibodies. Their three-complementarity determining region (CDR) loops contain all the necessary biochemical features to achieve nano-molar binding affinity to a given antigen, and have superior qualities for many applications relative to IgG’s, due to their smaller size and stability. Nanobodies have been employed effectively to trap transient conformations of medically relevant proteins for structural biology, facilitate non-invasive diagnostic imaging, imaging of dynamic processes in the cell, super resolution imaging of protein complexes, point of care diagnostic biosensors and as next generation cancer and SARS-CoV-2 therapies.


