Detection of fungal pathogens and their associated microbiome
Fungal pathogens are a major threat to human health, food production in agriculture, and biodiversity. In this project we aim to investigate the phyllosphere microbiome of wheat infected with different fungal pathogens. Your involvement in the project could be many fold from field sampling to bioinformatic analysis.
Project status
Content navigation
About
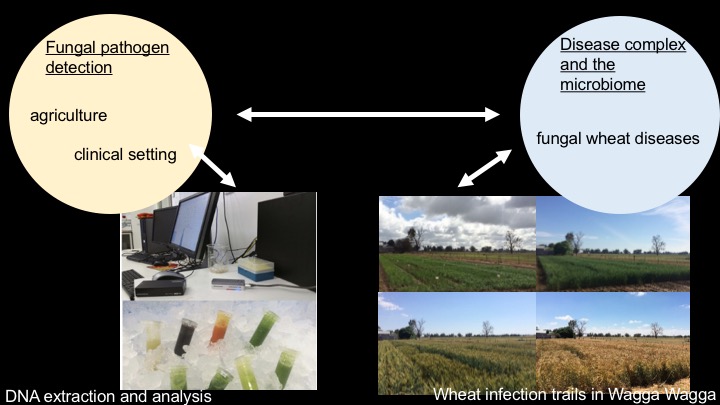
Fungal pathogens are a major threat to human health, food production in agriculture, and biodiversity (see Nature and Science). In this project we aim to develop novel detection protocols for fungi and their co-occurring microbial communities using a handheld DNA sequencer called the MinION from Oxford Nanopore. During the first stage of the project, we designed a series of experiments that enables us to develop best practices and suitable protocols for 'close to field detection' in the next stage of the project. Our current set-up includes targeted and whole genome sequencing of individual fungal species, mock communities, field samples from wheat, and human samples from diagnosed patients.
Your involvement in the project could be many fold from field sampling to bioinformatic analysis and pipeline development. This will depend on your level of interest and expertise.
Overall we aim to provide a fully-rounded training environment that exposes you to many aspects of modern biology such as experimental design, reproducible workflows, data management, data handling, visualization, and presentation.
This project is open to Honours, Master, and Summer Students mid-2020 and early-2021.

