A synthetic biology approach to detect pathogen molecules in crops
This project aims to develop an innovative rapid detection assay for pathogen molecules in wheat cells containing specific resistance genes. This assay will enable us to harness the full impact of the genomic revolution on plant pathology.
Research themes
Project status
Content navigation
About
Australian agriculture is a key contributor to the economy with the goal to grow the agricultural sector to AUD$100 billion within the next decade. Wheat is the major crop with a collective market value over AUD$6 billion. Fungal crop diseases are one of the major limitations causing yield losses of about 20% despite regular fungicide applications costing over $100 million a year. Natural genetic resistance is the preferred method of disease control as it is far more environmentally friendly than the application of hazardous chemicals. We have detailed molecular markers for many wheat resistance genes, which enables breeders to track these genes in wheat breeding programs and combine them in the desired configuration. In contrast, we know very little about which pathogen gene is recognised by each wheat resistance gene. This lack of knowledge leads to significant costs in practical disease management and laborious pathogen characterisation. We will address this issue by developing an innovative rapid detection assay for pathogen molecules in wheat cells containing specific resistance genes. This assay will enable us to harness the full impact of the genomic revolution on plant pathology. It will bridge the gap between bioinformatic prediction of candidate immunogenic pathogen genes and functional validation of recognition in wheat cells. The knowledge of many immunogenic pathogen genes will allow us to track them via cheap DNA sequencing and better predict which pathogen can infect what wheat variety.
The main objective of this proposal is to establish a highly innovative avirulence detector assay (ADA) in wheat. The three key attributes of ADA are; it is robust, with fast turnaround, and high-throughput. We will build on our experience with wheat protoplasts (intact plant cells lacking cell walls) which are readily obtained from 2-week-old wheat seedlings regardless of genetic background. This will make the assay broadly applicable as we will be able to test R genes in any wheat variety.
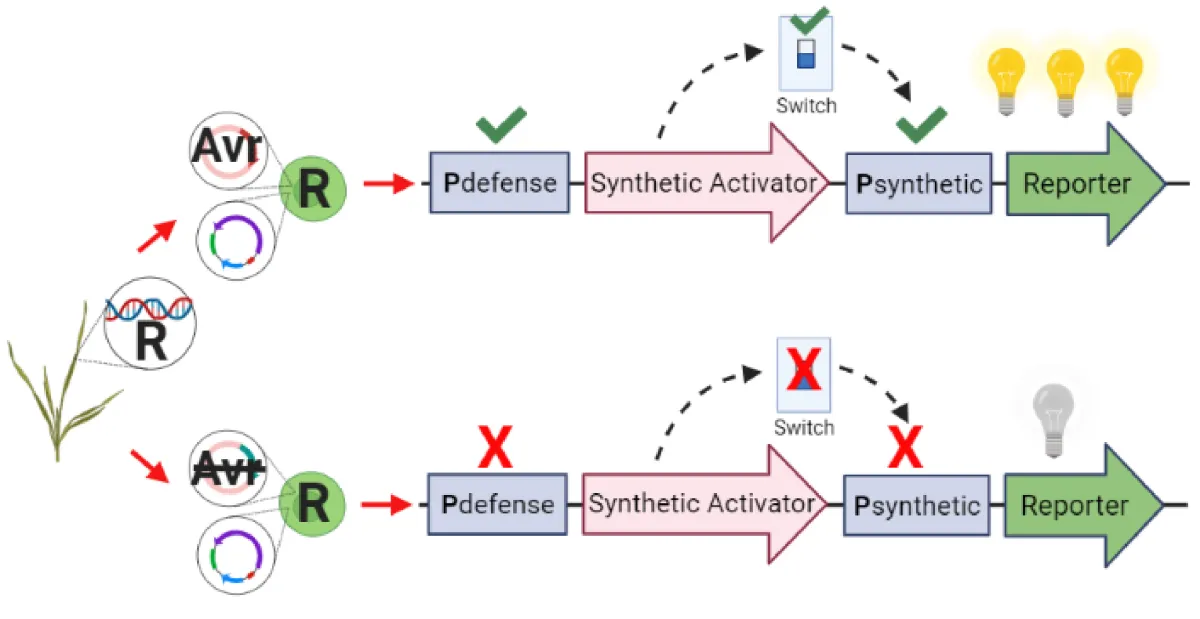
We will design a switch-like detector in which Avr/R recognition will drive the expression of a reporter (e.g. fluorescent protein) that can be selected on in a high-throughput automatic manner (e.g. fluorescence-activated cell sorting [FACS]). The protoplasts will be derived from a wheat variety with a known R gene and then transfected with DNA plasmids containing multiple different avirulence gene candidates in bulk. True Avr/R combinations will fluoresce and be captured. The DNA of the positive sorted cells will be extracted and sequenced to identify which candidate Avr triggered the immune response by the given natural R gene. This whole assay will take less than a week once established.
ADA relies on the fact that different Avr/R recognition events induce the expression of a common set of immune related genes. We will identify these genes in wheat protoplasts using three known Avr/R gene pairs in high-throughput gene expression assays (RNAseq). We will use the promoters of commonly induced genes to drive the expression of a reporter (e.g. GFP). We will combine this idea with the latest synthetic biology tools to generate a switch-like detector that fluoresces positive Avr/R protoplasts robustly and efficiently.

