Polyploidy and adaptation in Australian burrowing frogs Neobatrachus
Speakers
Event series
Content navigation
Description
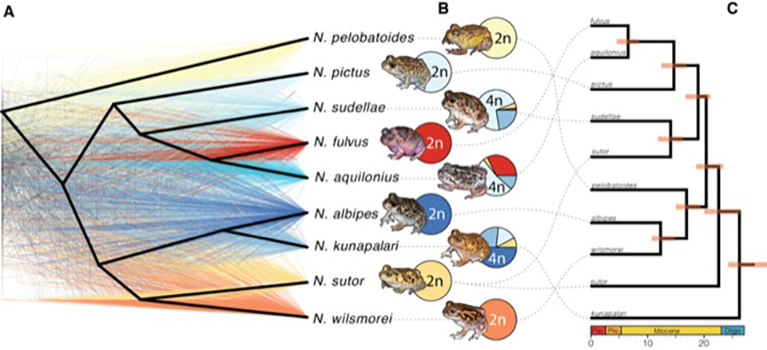
Polyploidy is rare in animals, and most polyploid animals reproduce asexually. Amphibians represent a dramatic vertebrate exception, with multiple independent sexually reproducing polyploid lineages. The Australian burrowing frog genus Neobatrachus is comprised of 6 diploid and 3 polyploid species and offers a powerful model animal polyploid system. We generated exome-capture sequence data from 87 individuals representing all 9 species of Neobatrachus to investigate population genomic effects of polyploidy on genus-wide demography. We document widespread gene flow between the tetraploids, asymmetric inter-ploidy gene flow directed from sympatric diploids to tetraploids, and current isolation of diploid species from each other. Changes in ecologically suitable areas corresponded to estimates of demographic histories and suggested that diploids may be suffering the early impacts of climate-induced habitat loss, while tetraploids appear to be avoiding this fate. At the same time, polyploids have to adapt their cellular machinery to ensure proper segregation of chromosomes during meiosis. We have assembled a draft genome of N. pictus using 10x Genomics technology and conducted a first selection scan between the N. pictus (2n) and the N. sudellae (4n). Our preliminary results show that selected genes in the tetraploids are enriched for microtubule motor activity function. This suggests modifications of the homologous pairing process during meiosis. Continuing this work we hope to provide the first description of adaptation mechanism(s) to autotetraploidy in animals. Overall, we demonstrate that Neobatrachus is an attractive model to study the effects of ploidy on evolution of adaptation in animals.
Location
Eucalyptus Seminar Room, Rm S205, Level 2, RN Robertson Building (46)
