E&E PhD Exit Webinar: Why Do Some Human Associated Escherichia coli Strains Lack Antibiotics Resistance?
The extra-intestinal pathogenic E. coli lineages STs 131, 73 and 95, of the phylogroup B2 are equally likely to be responsible for hospital- and community-acquired human extra-intestinal disease.
Speakers
Event series
Content navigation
Description
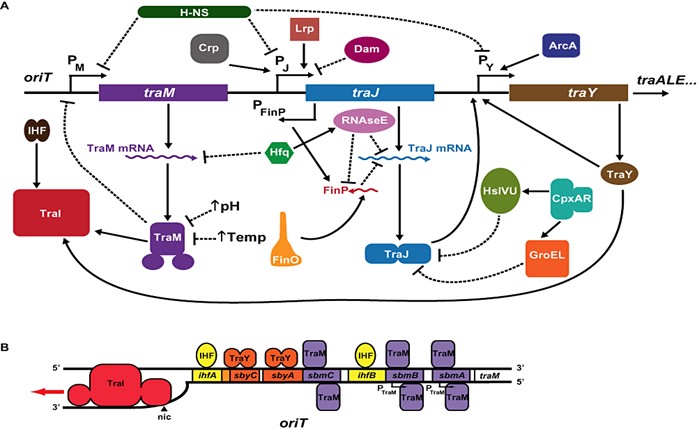
The extra-intestinal pathogenic E. coli lineages STs 131, 73 and 95, of the phylogroup B2 are equally likely to be responsible for hospital- and community-acquired human extra-intestinal disease. Given how ubiquitous these lineages are among humans, it would be expected that, in a community setting, the three lineages would be exposed to antimicrobials at about the same rate. However, many studies have demonstrated that their antimicrobial profiles differ: ST131 is noted to be highly resistant to antibiotics (Kallonen et al., 2017; Johnson et al., 2016), when compared with the other two lineages, STs 73 and 95. These later lineages have been observed to be largely antibiotics susceptible (Kallonen et al., 2017; Stephens et al., 2017; Gordon et al., 2017). The reasons for the antibiotic susceptibility of these extra-intestinal E. coli strains remains largely unknown. Given the genes conferring resistance to antimicrobials are frequently plasmid-borne, understanding the characteristics of the plasmids found in these STs in relation to their evolution and dissemination presents a key factor in unveiling what goes on within the plasmids of these lineages.
My doctoral thesis involved investigating the evolution, population dynamics, function, and molecular epidemiology of the plasmids within these lineages. I characterised the differences in the genomic diversity of the plasmids in the phylogroup B2 lineages using various bioinformatics tools. I investigated the plasmid – host dynamics by examining the rate of plasmid transfers among the ST lineages using plasmid transfer experiments, analysing the impacts of the plasmids on transfer rates using genome wide association studies, assessing how well the plasmids are maintained within the host after several generations using stability experiments and the fitness costs of the plasmids carriage within the host over time using competition experiments. Finally, I examined and compared the evolutionary history existing among the E. coli plasmids in the three lineages by using genome alignments techniques and phylogenetic analysis. The results of this study improve our understanding of the adaptation and evolution of plasmids of E. coli lineages.
Location
Please note: this seminar will be held in the Eucalyptus Rm and via Zoom, details are included below.
Eucalyptus Room, Rm S205, Level 2, RN Robertson Building (46)
Please click the link below to join the webinar: https://anu.zoom.us/j/84067505850?pwd=ZUZXaUJnbmExbGFwRTAwbGs2Njc1UT09
Passcode: 321465
Canberra time: please check your local time & date if you are watching from elsewhere.

